How species arise: a mathematical answer
Predicting when and how species arise is now possible with a new theoretical model using genome-wide data, developed by SIB and University of Bern researcher Simon Aeschbacher and colleagues. The study was published yesterday in the Proceedings of the National Academy of Sciences.
The diversity of life on our planet is stunning. Recent estimates indicate that there are several millions of species, and this number does not even include some of the most diverse groups of organisms, such as bacteria or viruses.
But how do species arise? Under what conditions will they persist? Can they arise without being spatially isolated?
"Answering these questions is challenging, as the process of speciation usually takes a long time compared to a researcher's lifespan," points out Simon Aeschbacher, an SIB scientist from the University of Bern. One way to solve the riddle is to observe, in vitro, speciation in organisms with very short generation times, such as viruses.
Another way, which is not restricted to a single species or organism, is to mine their genomes in silico, using mathematical models. Simon notes that "In theory, genome sequences are windows to the past. Mathematical models and bioinformatics tools can help us to interpret the information embedded in the genome of any organism at the level of species or populations." In a new study, published in the Proceedings of the National Academy of Sciences (PNAS), Simon and colleagues from the University of California and Duke University (USA) developed the first comprehensive framework for predicting when and how species arise, based on the genome-wide pattern of differentiation between two groups of organisms.
For the first time, this model is able to jointly estimate the strength of the two main opposing forces involved in speciation, as well as the timescale at which speciation is happening.
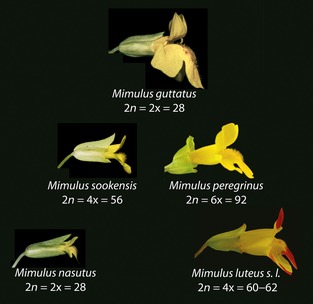
The team applied their framework to a real-world situation: how can two sister species of the monkeyflower Mimulus, living in the same geographical area, remain two distinct species in spite of sustained cross-pollination?
250,000-year-old "tug of war"
Speciation can be seen as a tug of war between two opposing forces. Gene flow, on the one hand, results from cross-mating between two populations. In the long run, it tends to prevent the formation of two separate species by leveling out their genetic differences. On the other hand, divergent selection renders hybrids resulting from such matings less ‘fit’ than their respective parents. So if selection is sufficiently strong relative to gene flow, the differentiation between the two populations will persist – which could eventually lead to speciation.
By applying their model to the yellow monkeyflower, M. guttatus, and its sister species M. nasutus, the team found that divergent selection has been engaged in a tug of war against gene flow for 250,000 years. This is what has kept these two species separate, in spite of strong levels of gene flow.
"It is impressive that we can estimate parameter values so small that they would remain undetected in direct observations", concludes Simon. "To gain the statistical power necessary for this strategy, information across the entire genome had to be aggregated."
The framework can be applied to an increasing number of species for which genome-wide data is becoming available. Researchers may therefore soon have a much better understanding of the mode and timing of speciation across a wide taxonomic range.
Publication details:Aeschbacher S, Selby J P, Willis J H and Coop G.: Population-genomic inference of the strength and timing of selection against gene flow. Proceedings of the National Academy of Sciences, 19 June 2017, doi: 10.1073/pnas.1616755114 |
Source: SIB
2017/06/27